Section: New Results
As-Rigid-As-Possible molecular interpolation paths
Participants : Krishna Kant Singh, Stephane Redon.
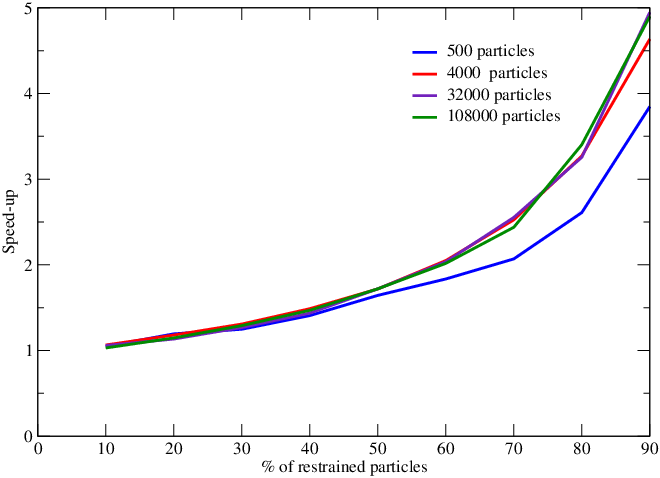
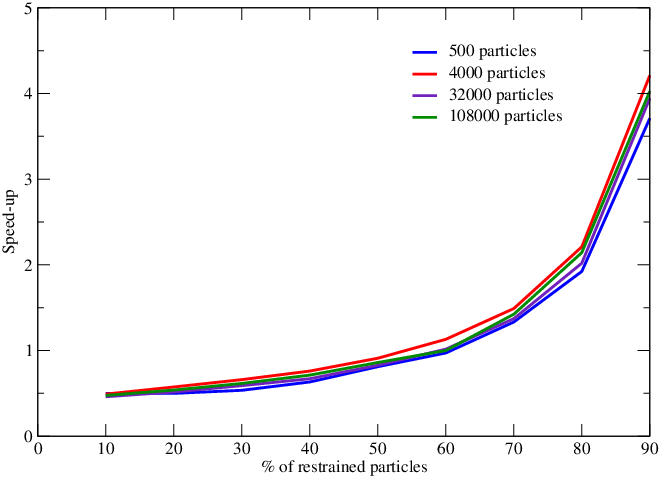
We have continued our work on the development of parallel adaptively restrained particle simulations. We proposed new algorithms to compute forces involving active particles faster. These algorithms involved construction of the Active Neighbor List (ANL) and incremental force computations. These algorithms have advantages over the state-of-the-art methods for simulating a system using Adaptively Restrained Molecular Dynamics (ARMD). Previously proposed algorithms required at-least restrained particles in order to achieve speed up. In new algorithms, we overcome this limitation and speed up can be achieve with retrained particles. We implemented our algorithm in the popular molecular dynamics package LAMMPS and submitted our results in the Computer Physics Communications Journal (K.K. Singh, S. Redon, Adaptively Restrained Molecular Dynamics in LAMMPS, Submitted to Computer Physics Communications.). Figure 11 show that speed-up can be achieved for more than of the particles are restrained. We also achieved significant speed up in constructing the ANL (figure 12).